RNA analysis
RNA sequencing
RNA sequencing is conducted in our partner company, when we get samples. Depending on species, polyA+RNA and/or RiboZero RNA are target of sequencing. As our standard, 40million reads are provided with 100bp or 150bp pair-end sequencing.
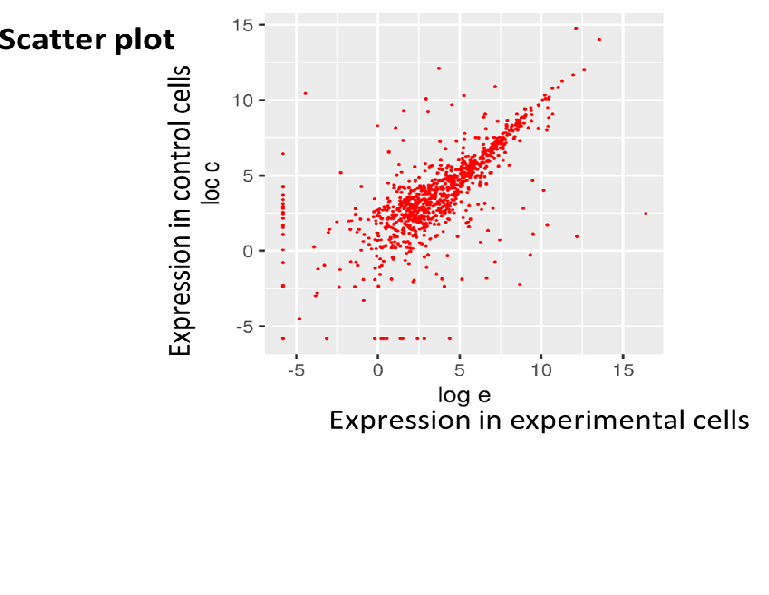
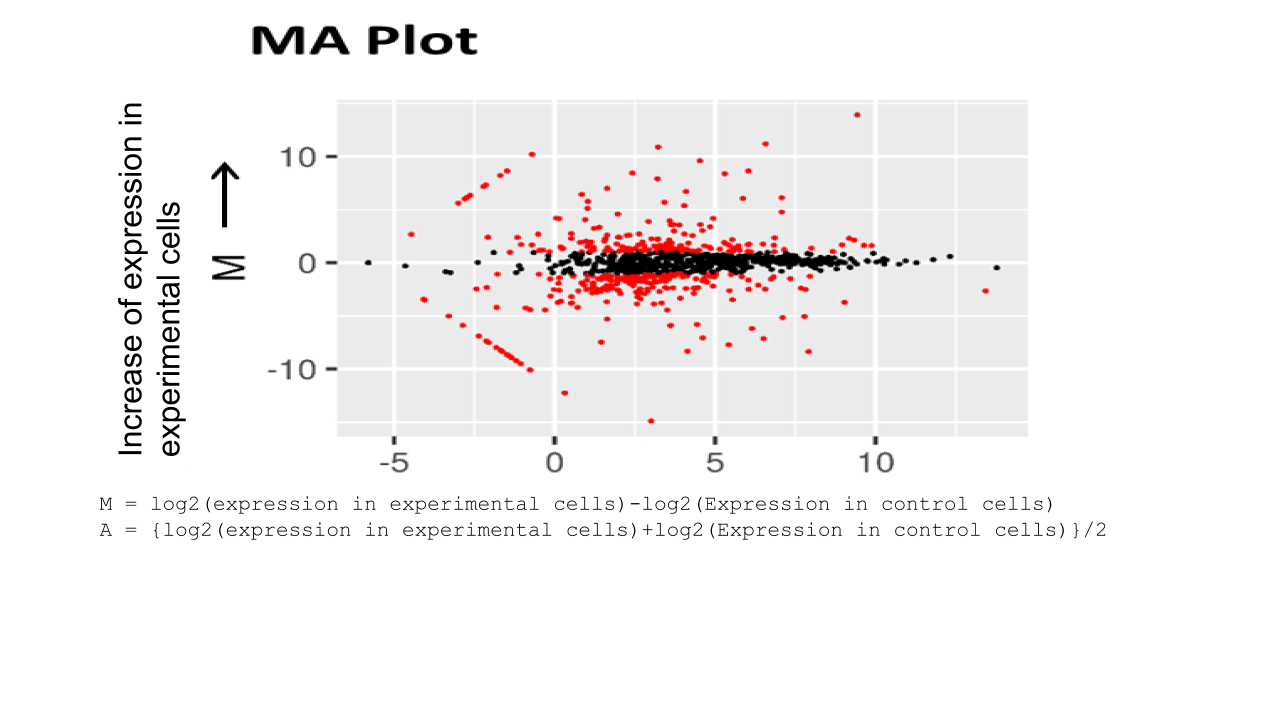
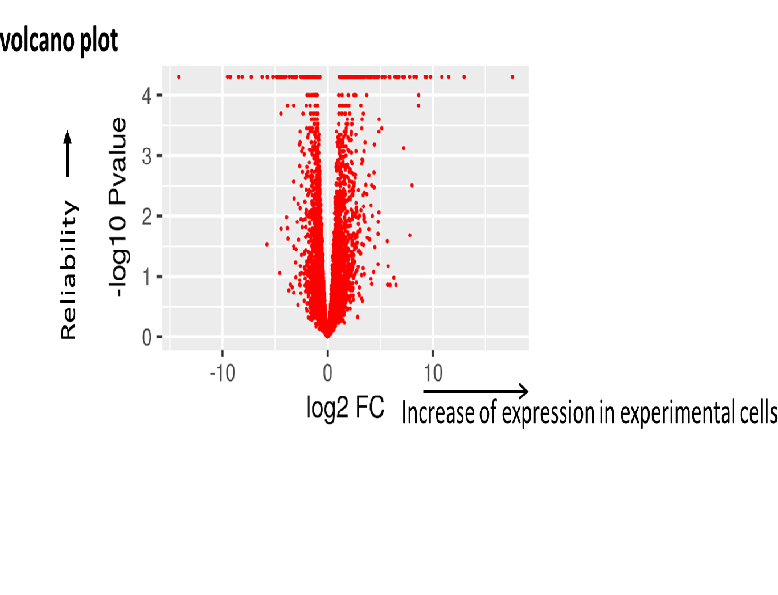
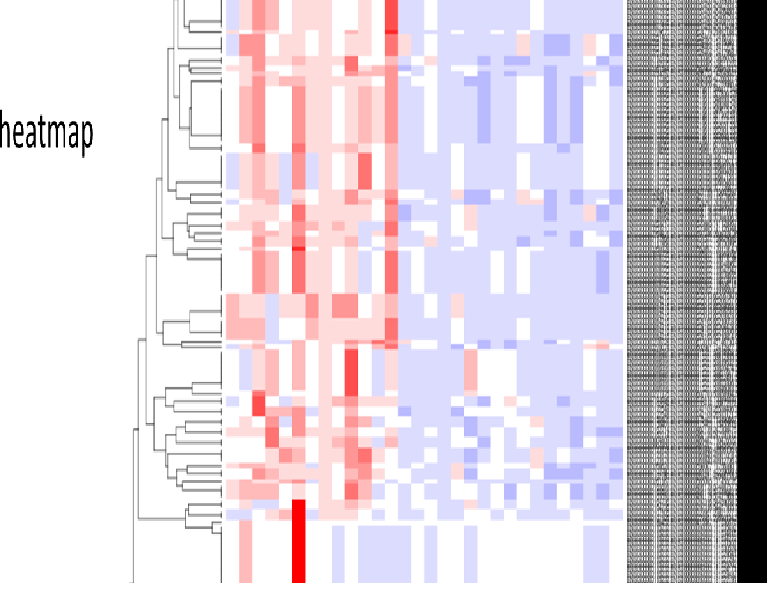
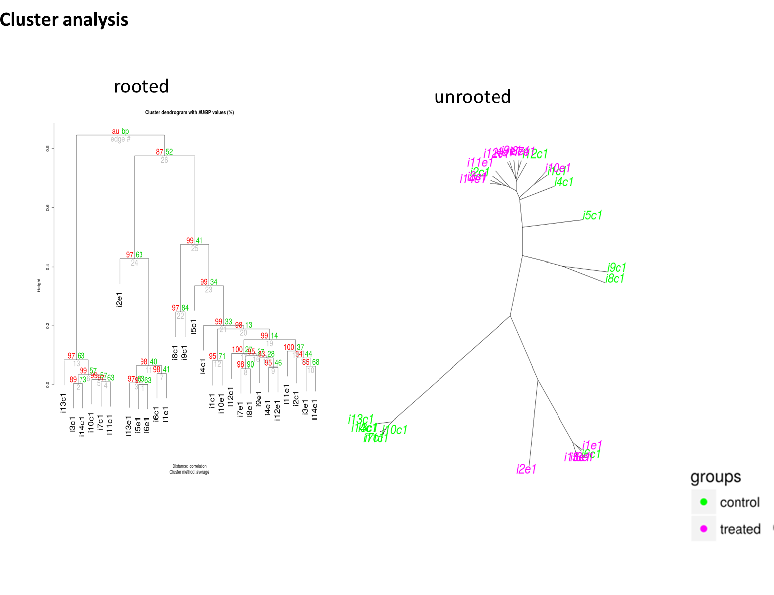
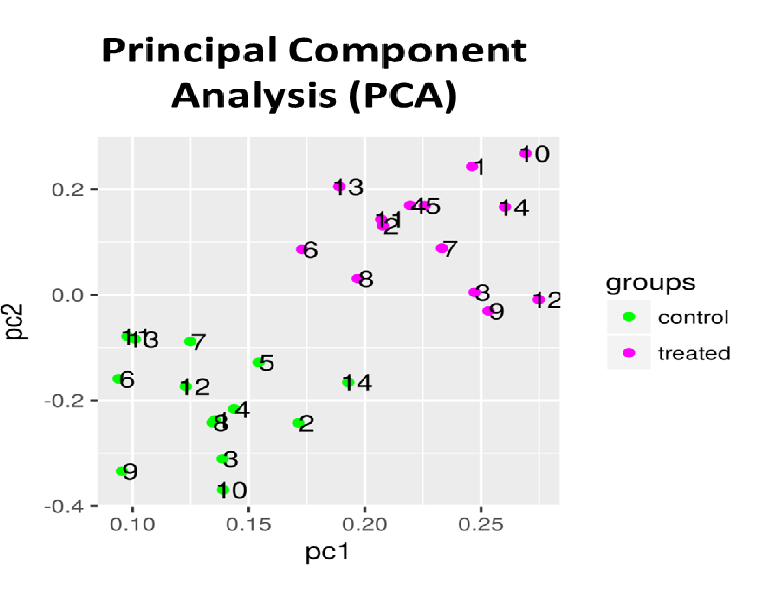
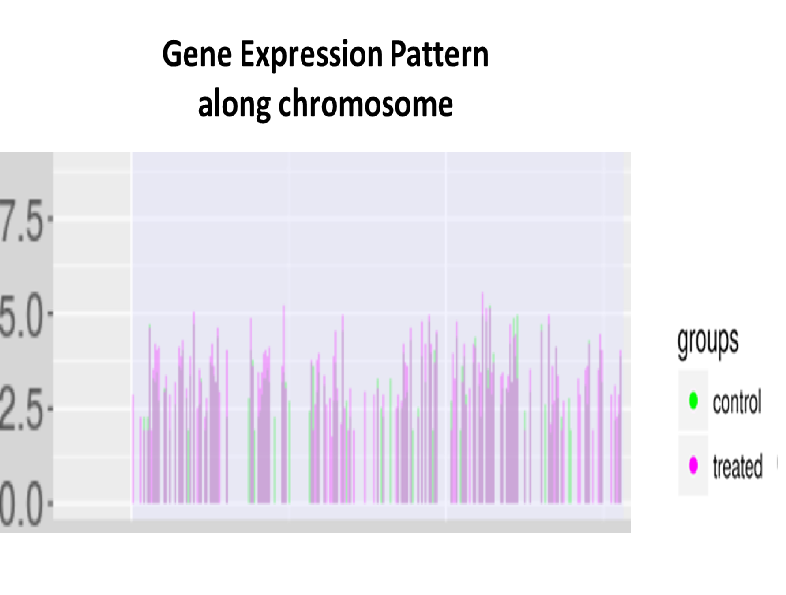
Expression analysis at gene and/or exon level
RNA sequence reads are first subjected to TopHat2 or STAR (at your choice) for mapping them to the latest reference genome, and aligned by Cufflinks. The resulting data are analyzed with Cuffdiff then with R libraries.
Lately, to obtain more concrete information about gene expression, in addition to in silico data, ‘wet examination’ is prerequisite using real-time PCR. We provide primer sequence information for the real-time PCR.
Primer design and real-time PCR
In order to confirm the in silico results in ‘wet experiments’, it is prerequisite to perform a real-time PCR using primer-pair designed inside of the one and same exon. In our analysis process, we provide the exon sequence and design a primer-pairs in the sequence.